What Is the Tumor Microenvironment?
The tumor microenvironment (TME) is the environment and community of cells surrounding a tumor. By releasing signals, tumors can control their microenvironment and its interaction with the body’s blood vessels, immune cells, and tissues and organs. These signals help cancer cells to spread and become resistant to both immune attack and medical intervention.
The number and kinds of cells that compose the TME vary with the type of cancer, stage, and treatment. They are thoroughly described here. For now, let’s think of a diverse population of cells from the tumor, immune system, vasculature (pericytes), extracellular matrix (fibroblasts, adipocytes), and lymphatic system. These cells interact to create a dynamic microenvironment known as the tumor microenvironment.
In this article, we'll briefly discuss TME niches (individual sub-micro environments within the TME). This includes their definition, creation, and impact within the greater tumor microenvironment. Our focus is on solid tumors, so the information shared might not apply to hematological malignancies.
Niches of the tumor microenvironment
"Broadly, six distinct specialized microenvironments with TME have been identified namely the hypoxic niche, acidic niche, innervated niche, metabolism microenvironment, immune microenvironment, and mechanical microenvironment."
Tiwari, A. et al.
Tumor microenvironment: barrier or opportunity towards effective cancer therapy →
Hypoxic niche
Growth of cancer cells leads to hypoxia, which stimulates expression of hypoxia inducible factor-1 (HIF-1), and ultimately stimulates vascular endothelial cells. HIF-1 induces angiogenesis, vascular infiltration of the tumor primarily via expression of VEGFA. HIF-1 also triggers expression of stem cell factors in cancer cells, leading to their further evolution along a survivalist path. The combination of HIF-1, VEGFA, and other hypoxia-induced genes correlate with a notably poor prognosis and decreased overall survival.
Acidic niche
Lactic acid buildup in the TME is initially toxic to all cells, including tumor cells. But cancer cells quickly adapt and preferentially utilize glycolysis. This maintains the acidic nature of the TME.
The acidic pH preferentially attracts tumorigenic immune cells including macrophages, neutrophils, and dendritic cells. Any anti-neoplastic immune cells entering this neighborhood are challenged by both hypoxia and low pH, hampering therapeutic interventions.
Innervated niche
When cancer invades a nerve or its route, called perineural invasion (PNI), the prognosis becomes quite grim. The cancer cells and nerves undertake a costimulatory cycle that leads to tumor growth and progression in a wide variety of cancer types.
This has recently become a very active area of research across a variety of tumor types. In a 2021 study, the Penn Ovarian Cancer Research Center explored the link between tumor innervation and poor prognosis. Increased nerve activity in the TME is linked in increased incidence and mortality for cancer patients. Nerve activity also helps cancer evade treatment that would otherwise be effective.
Research points to the nervous system as an important target for cancer treatment. Treatment strategies in development include denervation, targeting axonogenic signaling, and antineurotrophic therapy. However, because the neural activity varies in different types of cancer, more research is necessary to understand tumor-nerve interaction.
Metabolism microenvironment
Between hypoxia and tumor cells that preferentially use glycolysis, the TME’s metabolic environment could best be described as inhospitable. Incoming immune cells, including those designed as targeted therapies, will find nutrients scarce (low vascularization), acidic pH (high lactate), and abundant reactive oxygen molecules damaging their DNA.
Immune microenvironment
The immune system consists of two arms, innate and adaptive. Components of both are attracted to the TME by signals secreted by tumor cells. Cells of the adaptive immune system, including T-lymphocytes, are recruited by the tumor but often encounter a highly immunosuppressive microenvironment. This in turn can lead to exhaustion of the invading cytotoxic-T cells and loss of their anti-tumor activity.
An immunosuppressive microenvironment recruits additional suppressive cells including regulatory T and B cells, and the bane of all immunotherapies, myeloid-derived suppressor cells (MDSCs).
Extensive study of the immune environment led Nobel Laureate Carl June to bioengineer killer T-cells to attack tumors. These chimeric antigen receptor -T cells (CAR-T) have shown great success in treating hematological malignancies. They have even delivered >10-years cancer-free to recipients like Emily Whitehead.
Targeting tumors with CAR-T cell therapies or by stimulating specific components of the immune system are relatively recent approaches in the fight against cancer. One of the major hurdles in CAR-T therapies is the timely manufacturing of these modified patient-derived cells.
CAR-T are generated by taking blood from patients, isolating the T-cells, activating them, and genetically modifying the cells to recognize the tumor. Those genetically engineered cells are then returned into patients. As we develop more efficient processes to manufacture CAR-T cells, pathologists are working hard to develop methods that more accurately predict who can benefit from specific therapeutic approaches.
One of the most exciting techniques being investigated by pathologists involves using AI to measure the distance between certain kinds of immune cells and their neighboring tumor cells to provide a more accurate prognosis. For example, Nucleai used a deep learning model to investigate how TME composition impacts clinical outcomes.
This program provided cell typing at higher accuracy than existing approaches. It also identified a link between plasma cells and prognosis in colorectal cancer. This model was successful across types of cancer and biomarkers.
Mechanical microenvironment
The role of mechanical stimulation is another highly active area in cancer research. Intracellular components like actin fibers and the extracellular matrix (ECM) of fibers, stromal cells, and their secreted factors can remodel the tumor’s environment and promote invasion. "Stiffness” of the ECM also contributes to cancer progression in some tumor types.
The ECM proteins collagen and elastin are primary contributors to stiffening. In combination with multiple signaling pathways, especially TGF-Beta, chaperones like Hsp47 and SPARC, deposition of collagen is increased along with tension in the ECM.
The increased mechanical stress on mammary epithelial cells from a tense ECM has been demonstrated to induce malignant transformation. This oncogenic effect is not limited to mammary tissues. It has also been seen in the pancreas, liver, and other tissues.
Researchers recognize that ECM is an important component to creating more effective treatments. Our development of approaches to modify ECM safely and effectively in conjunction with existing therapeutics is another critical area of cancer research.
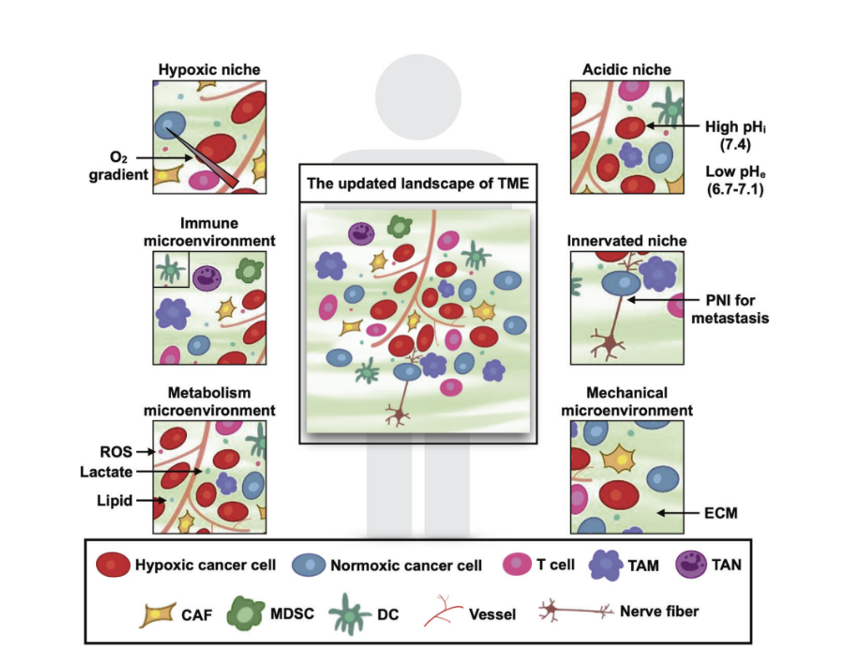 Jin, MZ., Jin, WL. The updated landscape of tumor microenvironment and drug repurposing. Sig Transduct Target Ther 5, 166 (2020).
Jin, MZ., Jin, WL. The updated landscape of tumor microenvironment and drug repurposing. Sig Transduct Target Ther 5, 166 (2020).
Creation of the Tumor Microenvironment
Researchers widely believe that the tumor microenvironment is created by the first tumor cells secreting signaling molecules. These molecules recruit a wide variety of immune and “normal” cell types to form the associated stroma. Causes of the neoplastic transformation of healthy cells to tumors is an exceedingly complex topic. Many hypotheses, known contributing factors, and potential mechanisms make it difficult develop certainties about this process.
Technological advancements such as single-cell sequencing and spatial transcriptomics are creating resources describing the gene expression programs of healthy cells and tissues. This includes projects like the Human Cell Atlas (HCA) and Human BioMolecular Atlas Program (HuBMAP). Researchers will now have a very early baseline defining “healthy gene expression” to monitor the progression to a diseased cell state.
Technological Advancements to Study the TME
Early reports on the TME focused on the contributions of tumor cells. These were considered to dominate the behavior of all cells in the area. Our understanding was limited by low resolution approaches such as whole genome sequencing and bulk RNA sequencing. Only the averages of gene expression changes were captured.
Subtle changes in gene expression, cell populations, and/or mutations were not detectable. In that historical context, it is not surprising that the presence of immune cells was reported to be both a poor and a good prognostic indicator.
Early methods to assess protein expression were also limited to analyzing bulk tissue homogenates by Western blot or single-plex immunohistochemistry (IHC) on tissue sections/biopsies. Historically, a key instruction when selecting tissues for clinical IHC was to ensure that the area on the slide contains >50% tumor by morphology.
Advancements in Single-Cell Technology
Our technology has advanced significantly since the early days when tumors and TMEs were homogenized for bulk sequencing. It is now possible to analyze the complete transcriptomes of single cells alone or dissociated from tissue sections. Single-cell sequencing is now used by many researchers to characterize the transcriptional programs of individual cells.
Until recently, this technology was limited to live cells and those dissociated from frozen tissues. New chemistries like the Flex Fixed Gene Expression permit transcriptome mapping of cells that are frozen in time-thanks to chemical fixation. These experiments lead to a wealth of new information.
Spatial Proteomics Applied to the TME
The paradigm for all pathology tissue samples to include the maximum amount of tumor was turned on its head by David Rimm’s pathology lab. In 2019, his team used the GeoMx Digital Spatial Profiler in TME research. Rimm’s data showed that the level of multiple proteins within the tumor stroma (not the tumor itself) correlated strongly with positive responses to immunotherapy in late-stage melanoma.
Today’s cutting-edge technology can detect transcripts within individual cells that are still within their native tissue context. Spatial biology instruments can map the levels and locations (X, Y, Z coordinates) of individual RNAs or proteins at subcellular resolution in tissues on slides. It is now possible to interrogate the cells of the TME with unprecedented resolution. Researchers can obtain that same deep level of information about the nearest neighboring cells to create a comprehensive map of the TME.
Therapeutic Implications of the TME
Scientists and clinicians now know that the immune system recognizes and infiltrates the tumor’s neighborhood. Therapeutic developments are attempting to capitalize upon this information. Researchers and clinicians are utilizing targeted therapies including chimeric antigen receptor (CAR-T), immune modulation via checkpoint inhibitors, and cancer vaccines. These treatments, either alone or in combination, treat a wide range of cancers, including solid tumors.
The dynamic interplay among all cell types in the surrounding tissue, especially cancer and immune cells, represents a much clearer picture of conditions within the TME. Spatial biology is only in its infancy, but it's clear this tool can revolutionize how we diagnose and treat cancer.
AI-driven tools to measure the distances between immune and tumor cells within the TME will increase the accuracy of prognosis. The incredible sensitivity of new technologies will expand our understanding of the tumor microenvironment and the fundamentals of cell biology.



